Health Data is becoming an increasing important source for clinical and genomic research. Researchers create and iteratively refine algorithms using structured and unstructured data to better identify cohorts of subjects within the health data.
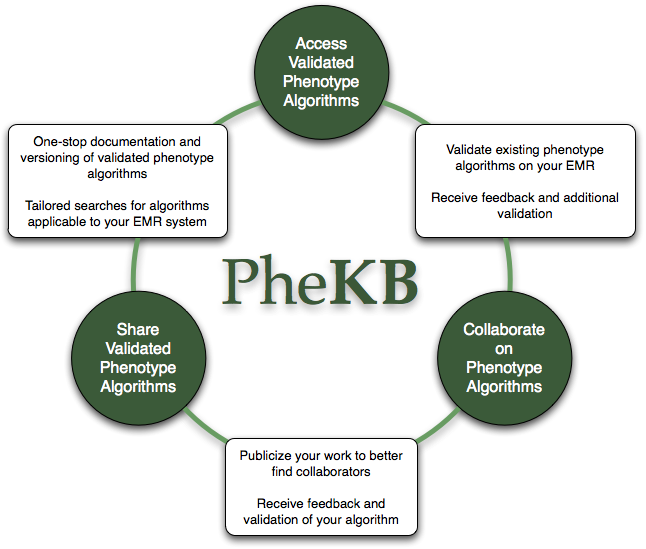
The Phenotype Knowledgebase website, PheKB, is a collaborative environment to building and validating electronic algorithms to identify characteristics of patients within health data. PheKB was functionally designed to enable such a workflow and has purposefully integrated tools and standards that guide the user in efficiently navigating each of these stages from early stage development to public sharing and reuse. PheKB has tools to enable cross-site collaboration for algorithm development, validation, and sharing for reuse with confidence.
On PheKB you can: View existing algorithms; Enter or create new algorithms; Collaborate with others to create or review algorithms; View implementation details for existing algorithms
Phenotype algorithms can be viewed by data modalities or methods used:
- ICD and CPT codes
- Laboratories
- Medications
- Vital Signs
- Natural Language Processing
PheKB Partners
Reference
Kirby JC, Speltz P, Rasmussen LV, Basford M, Gottesman O, Peissig PL, Pacheco JA, Tromp G, Pathak J, Carrell DS, Ellis SB, Lingren T, Thompson WK, Savova G, Haines J, Roden DM, Harris PA, Denny JC. PheKB: a catalog and workflow for creating electronic phenotype algorithms for transportability. J Am Med Inform Assoc. 2016 Mar 28; PMID: 27026615



